Mindy Melinda A Engevik PhD
Faculty email addresses should not be used to seek medical advice or to make medical appointments. Please visit
MyChart for medical appointments or to contact your provider.
Biography
Dr. Engevik received her PhD from the University of Cincinnati, and joined the MUSC faculty in 2020 following postdoctoral work studying the interaction of microbial metabolites and the gut epithelium with a focus on inflammatory bowel disease at Baylor College of Medicine in Houston, TX.
Dr. Engevik has pursued a lifetime interest in the interactions between microbes and epithelial cells of the gut. Her past graduate work included studies of immune system responses to Candida albicans fungus, mechanisms regulating ammonia transport in the colon, and ion transport-mediated regulation of the intestinal environment and gut microbiota by gut epithelial cells. She continued to study the interaction between microbial metabolites and the gut epithelium during her postdoctoral work. This postdoctoral work included pioneering studies on the utility of human intestinal organoids, also known as enteroids or colonoids, derived from tissue-derived stem cells for modeling microbe-host interactions.
Dr. Engevik’s current research continues to focus on microbial-host crosstalk, now with an emphasis on interactions between microbes and the intestinal mucus layer. This focus encompasses the two-sided nature of these interactions: the beneficial effects on host health mediated by commensal microbe modulation of the mucus layer, and the subversion of mucus function during infection by colonizing pathogens. These efforts are facilitated by the availability of both animal and human organoid model systems through the DDRCC and CDLD cores.
Our recent understanding of the gut microbiota has sparked interest
in investigating microbes’ ability to colonize and modulate the gut
environment. The intestinal mucus layer serves as the first point of
contact between the gut microbiota and the host. In addition to
providing a barrier for the epithelium, the mucus layer provides a niche
for bacteria due to its abundance of attachment sites and potential as a
nutrient source. Dr. Mindy Engevik’s lab investigates microbial-host crosstalk with an emphasis on microbe-mucus interactions.
The Engevik lab has 2 main focuses: (1) how commensal microbes
beneficially modulate the mucus layer and host health; and (2) how
pathogens colonize and subvert the mucus layer to cause infection.
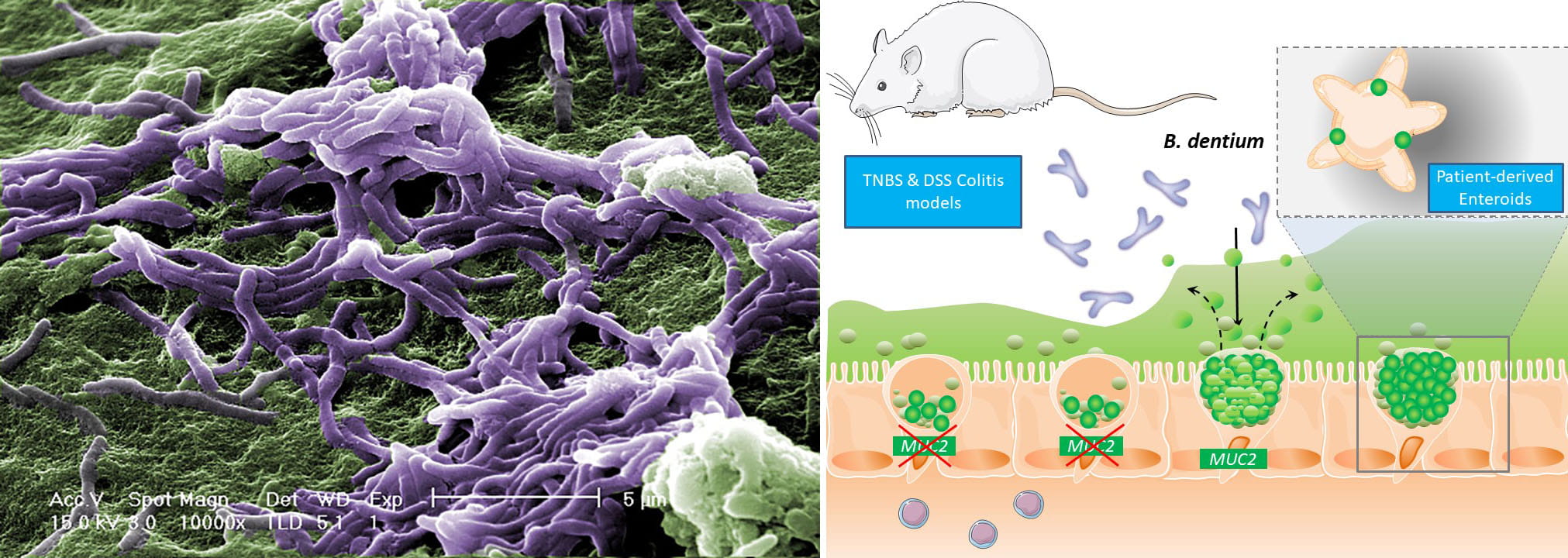
(1) One of our favorite commensal microbes is Bifidobacterium dentium.
We have identified that B. dentium metabolites stimulate the release of
serotonin from intestinal enterochromaffin cells. Serotonin activates up
to 14 different intestinal serotonin receptor subtypes, including ones
on mucus-producing goblet cells. We hypothesize that serotonin activates
goblet cell 5-HTR4, stimulating the release of mucus and the healing
peptide trefoil factor (TFF3). Our preliminary data with human
intestinal organoid, also known as enteroid, monolayers indicates that
serotonin and B. dentium metabolites enhance wound healing. So now we
need to dissect the mechanisms, identify candidate metabolites, examine
these effects in vivo and more! Ultimately, these studies could provide
new targets for promoting epithelial repair and wound healing.
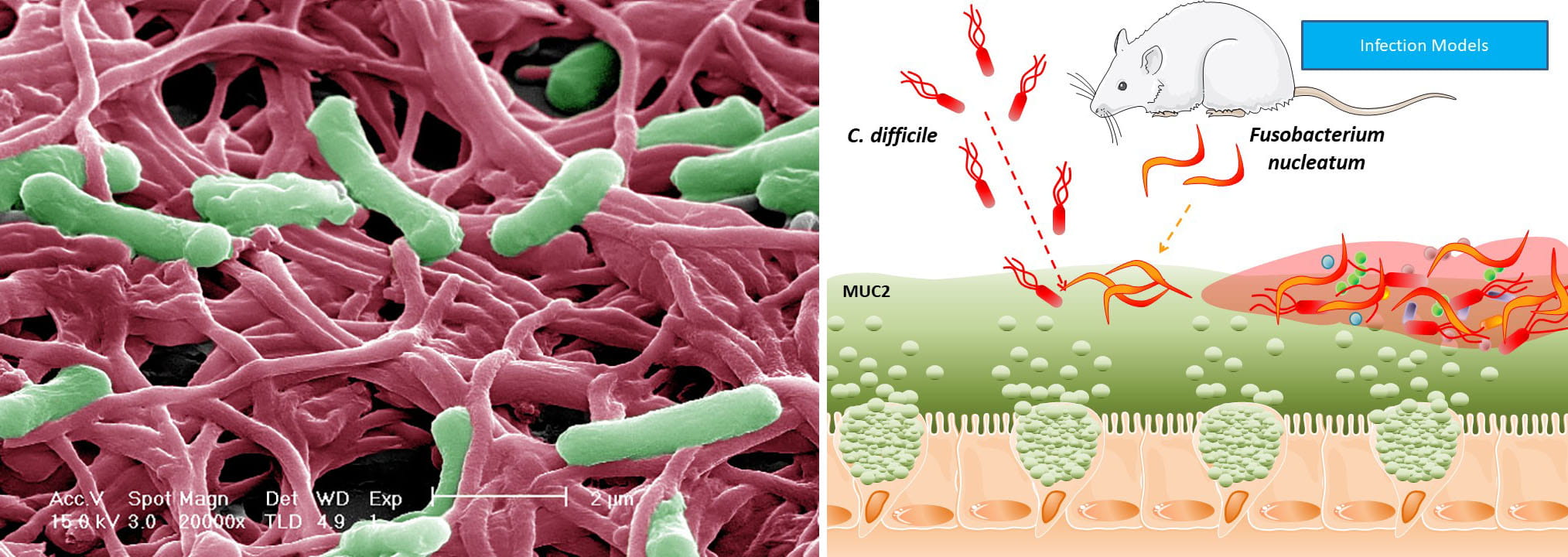
(2) In terms of pathogens, we also study Clostridium difficile. C.
difficile which is the most common healthcare associated pathogen in
U.S. hospitals, incurring billions of dollars in treatment costs each
year. Studies have shown that antibiotic disruption of the gut
microbiota creates a favorable niche for C. difficile spore germination
and ultimately toxin production and disease. Our previous work has
demonstrated that C. difficile adheres to intestinal mucus with
mucin-degrading bacteria such as Bacteroides, Ruminococcus, and
Akkermansia. We also identified a high abundance of Fusobacterium, an
oral microbe known to form multi-species biofilms. We now want to know
how Fusobacterium and these other microbes influences C. difficile
infection. These studies will characterize microbe-microbe signaling
networks involved in C. difficile infection.